Darin Tsui

Hi there! My name is Darin. I’m a Ph.D. candidate and NSF GRFP fellow at the Georgia Institute of Technology studying Electrical and Computer Engineering and Bioengineering. I previously received my BS degree at the University of California San Diego in Bioengineering.
I currently work with Dr. Amirali Aghazadeh in the AI ML & Information Group, where I develop scalable, interpretable tools for generative models in the biosciences. I am currently supported by the NSF Graduate Research Fellowship. My research focuses on:
-
Explaining black-box models at scale by designing algorithms at the intersection of signal processing, coding theory, and interpretabiity.
-
Leveraging mechanistic interpretability to extract higher-order interactions and discover actionable biological knowledge.
-
Developing principled generative models that leverage domain priors.
My background is highlighly interdisciplinary. I previously validated deep learning models at Surgalign (acquired by Xtant Medical), developed an optical surgical platform in the Talke Biomedical Device Lab, and designed a bioelectronic COVID-19 test in the Integrated Systems Neuroengineering Laboratory.
Outside of research, I co-chair Bioengineering Day at Georgia Tech, which aims to highlight interdisciplinary research. Previously, I served as President of the IEEE student branch and helped found the IEEE EMBS chapter at UC San Diego.
News
| Oct 02, 2025 | I gave a guest lecture in ECE 8803–Generative and Geometric Deep Learning–on explainability and mechanistic interpretability in AI. You can find the recording of the lecture here. |
|---|---|
| Sep 24, 2025 | My work on adapting sparse autoencoders for low-N protein engineering into the NeurIPS AI4Science workshop. |
| Sep 18, 2025 | My work on explaining biological sequence models at scale has been accepted into NeurIPS 2025! |
| Sep 18, 2025 | Our work on adapting speculative decoding for protein generation was accepted to NeurIPS 2025 as a spotlight! |
| Jul 16, 2025 | Our work on assessing and explaining mutations in myocilin was accepted to the Machine Learning in Computational Biology (MLCB) meeting. |
Selected Publications
-
 SpecMER: Fast protein generation with K-mer guided speculative decodingIn NeurIPS (Spotlight) , 2025
SpecMER: Fast protein generation with K-mer guided speculative decodingIn NeurIPS (Spotlight) , 2025 -
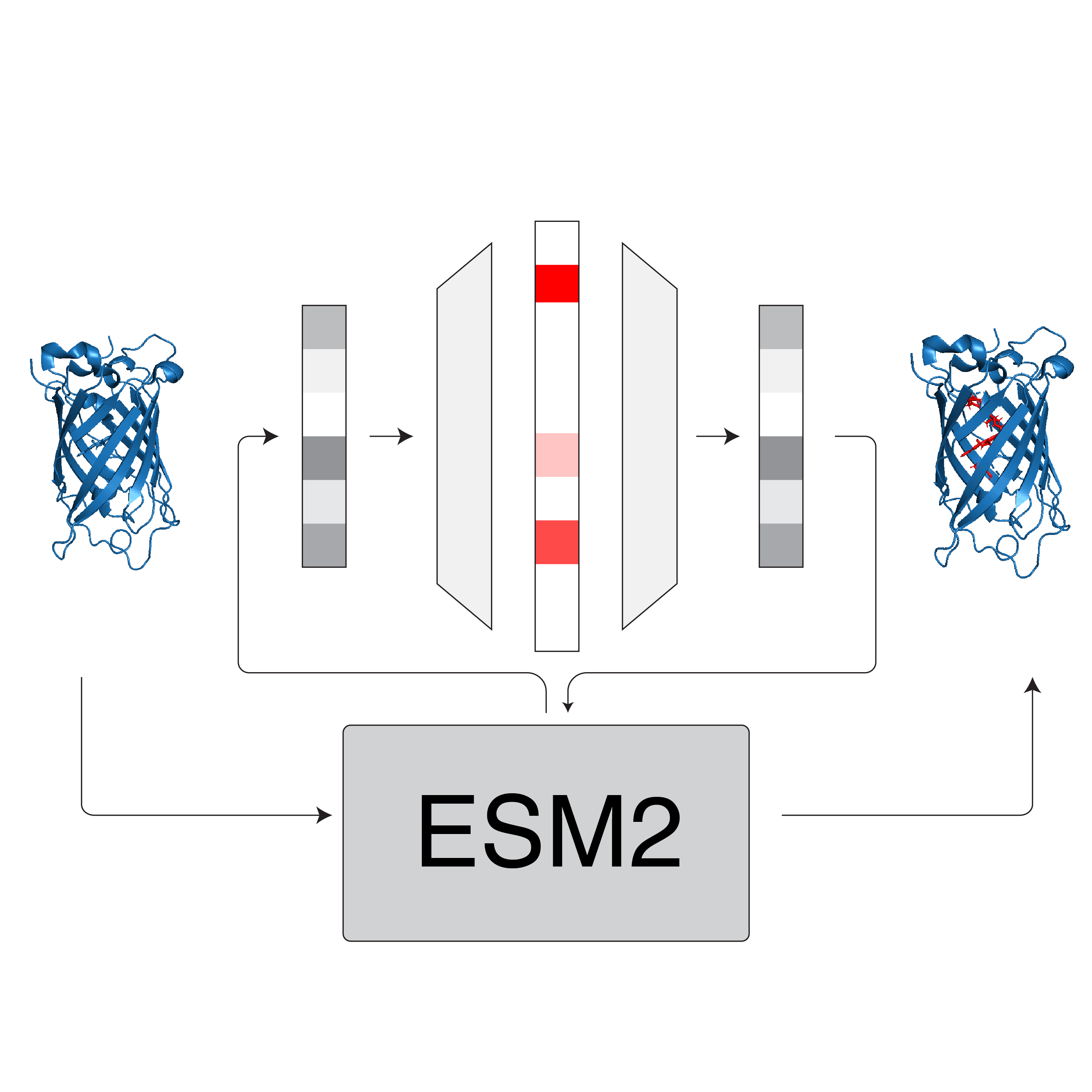 Sparse Autoencoders for Low-N Protein Function Prediction and DesignIn NeurIPS AI4Science Workshop , 2025
Sparse Autoencoders for Low-N Protein Function Prediction and DesignIn NeurIPS AI4Science Workshop , 2025
